Tutorial: Partition your space
Why partitioning the space?
Often the data is not written on disks the way we would like to have it distributed. Re-partitioning the space is a way to re-organise the data to speed-up later queries, exploration of the data set or visualisation.
Unfortunately, re-partitioning the space involves potentially large shuffle between executors which have to send and receive data according to the new partitioning scheme. Depending on the network capacity and the size of the initial data set, this operation can be costly and the re-partition is only a good idea if multiple queries have to be performed, or if you do not have other choices (like in visualisation).
You might noticed the DataFrame API does not expose many things to repartition your data as you would like (unlike the RDD API), hence we decided to bring here new DataFrame features!
How spark3D partitions DataFrames?
The partitioning of a DataFrame is done in two steps.
- First we create a custom partitioner, that is an object which associates each object in the dataset with a partition number. Once this partitioner is known, we add a DataFrame column with the new partition ID (
prePartition). - Second we trigger the partitioning based on this new column (
repartitionByCol) using a simpleKeyPartitioner.
Note that partitioning by a DataFrame column is similar to indexing a column in a relational database.
Available partitioning and partitioners
There are currently 2 spatial partitioning implemented in the library:
- Onion Partitioning: See here for a description. This is mostly intented for processing astrophysical data as it partitions the space in 3D shells along the radial axis, with the possibility of projecting into 2D shells.
- Octree: An octree extends a quadtree by using three orthogonal splitting planes to subdivide a tile into eight children. Like quadtrees, 3D Tiles allows variations to octrees such as non-uniform subdivision, tight bounding volumes, and overlapping children.
In addition, there is a simple KeyPartitioner available in case you bring your own partitioning scheme (see example below).
An example: Octree partitioning
In the following example, we load 3D data, and we re-partition it with the octree partitioning (note that in practce you can have metadata in addition to the 3 coordinates):
from pyspark3d.repartitioning import prePartition
from pyspark3d.repartitioning import repartitionByCol
# Load data
df = spark.read.format("csv")\
.option("header", True)\
.option("inferSchema", True)\
.load("src/test/resources/cartesian_spheres_manual.csv")
df.show()
+---+---+---+------+
| x| y| z|radius|
+---+---+---+------+
|1.0|1.0|1.0| 0.8|
|1.0|1.0|1.0| 0.5|
|3.0|1.0|1.0| 0.8|
|3.0|1.0|1.0| 0.5|
|3.0|3.0|1.0| 0.8|
|3.0|3.0|1.0| 0.5|
|1.0|3.0|1.0| 0.8|
|1.0|3.0|1.0| 0.5|
|1.0|1.0|3.0| 0.8|
|1.0|1.0|3.0| 0.5|
|3.0|1.0|3.0| 0.8|
|3.0|1.0|3.0| 0.5|
|3.0|3.0|3.0| 0.8|
|3.0|3.0|3.0| 0.5|
|1.0|3.0|3.0| 0.8|
|2.0|2.0|2.0| 2.0|
+---+---+---+------+
# Specify options.
options = {
"geometry": "points",
"colnames": "x,y,z",
"coordSys": "cartesian",
"gridtype": "octree"}
# Add a column containing the future partition ID.
# Note that no shuffle has been done yet.
df_colid = prePartition(df, options, numPartitions=8)
df_colid.show()
+---+---+---+------+------------+
| x| y| z|radius|partition_id|
+---+---+---+------+------------+
|1.0|1.0|1.0| 0.8| 6|
|1.0|1.0|1.0| 0.5| 6|
|3.0|1.0|1.0| 0.8| 7|
|3.0|1.0|1.0| 0.5| 7|
|3.0|3.0|1.0| 0.8| 5|
|3.0|3.0|1.0| 0.5| 5|
|1.0|3.0|1.0| 0.8| 4|
|1.0|3.0|1.0| 0.5| 4|
|1.0|1.0|3.0| 0.8| 2|
|1.0|1.0|3.0| 0.5| 2|
|3.0|1.0|3.0| 0.8| 3|
|3.0|1.0|3.0| 0.5| 3|
|3.0|3.0|3.0| 0.8| 1|
|3.0|3.0|3.0| 0.5| 1|
|1.0|3.0|3.0| 0.8| 0|
|2.0|2.0|2.0| 2.0| 1|
+---+---+---+------+------------+
# Trigger the repartition
df_repart = repartitionByCol(df_colid, "partition_id", preLabeled=True, numPartitions=8)
df_repart.show()
+---+---+---+------+------------+
| x| y| z|radius|partition_id|
+---+---+---+------+------------+
|1.0|3.0|3.0| 0.8| 0|
|3.0|3.0|3.0| 0.8| 1|
|3.0|3.0|3.0| 0.5| 1|
|2.0|2.0|2.0| 2.0| 1|
|1.0|1.0|3.0| 0.8| 2|
|1.0|1.0|3.0| 0.5| 2|
|3.0|1.0|3.0| 0.8| 3|
|3.0|1.0|3.0| 0.5| 3|
|1.0|3.0|1.0| 0.8| 4|
|1.0|3.0|1.0| 0.5| 4|
|3.0|3.0|1.0| 0.8| 5|
|3.0|3.0|1.0| 0.5| 5|
|1.0|1.0|1.0| 0.8| 6|
|1.0|1.0|1.0| 0.5| 6|
|3.0|1.0|1.0| 0.8| 7|
|3.0|1.0|1.0| 0.5| 7|
+---+---+---+------+------------+
preLabeled means the column containing the partition ID contains already numbers from 0 to numPartitions - 1 (see below for the generic case).
Options for 3D repartitioning
The current options for spark3D repartitioning are:
gridtype: the type of repartitioning. Available: current (no repartitioning), onion, octree.geometry: geometry of objects: points (3 coordinates), spheres (3 coordinates + radius)coordSys: coordinate system: spherical or cartesiancolnames: comma-separated names of the spatial coordinates. For points, must be “x,y,z” or “r,theta,phi”. For spheres, must be “x,y,z,R” or “r,theta,phi,R”.
For example, to load point-like objects whose column coordinates are (x, y, z), in a cartesian coordinate system and repartition using octree, you would use:
options = {
"geometry": "points",
"colnames": "x,y,z",
"coordSys": "cartesian",
"gridtype": "octree"}
Create your own Partitioning
spark3D contains limited number of partitioners. We plan to add more (RTree in preparation for example). In the meantime, you can add your partitioner, and use spark3D repartitionByCol to trigger the repartitioning.
Typically, a partitioner is an object which associate to each row a partition ID. So if you come up with your column containing partition ID, the job is done!
Here is a simple repartitioning. Let’s suppose we have a DataFrame with 4 columns (x, y, z, color):
df.show(5)
+-------------------+-------------------+--------------------+-----+
| x| y| z|color|
+-------------------+-------------------+--------------------+-----+
| 0.7597103249274305|0.43553591641478184| 0.28917234457456076| C0|
| 0.7026135458509866| 0.8153555174579074| 0.2271205402627411| C1|
|0.29406270599653683| 0.1279417475009852| 0.22566980128052516| C0|
| 0.9243776892995804| 0.4338940983421623| 0.46862943234999066| C1|
|0.13688472232767646| 0.517193173845641| 0.2571428515204228| C0|
| 0.5546555520340469| 0.9007788514727354|0.023848244237575034| C1|
| 0.3020298950179151| 0.4970200882013287| 0.5753172391955962| C0|
| 0.7973444219353029| 0.78955164333523| 0.06861238579818774| C1|
| 0.9970636495720013|0.46611372809037144| 0.18037269668312206| C0|
| 0.2656533532231762| 0.8281379193317686| 0.13374486702332633| C1|
+-------------------+-------------------+--------------------+-----+
We will create a column whose elements are based on the distance of (x, y, z) to the center (0, 0, 0):
from pyspark.sql.functions import pandas_udf
from pyspark.sql.functions import PandasUDFType
@pandas_udf("double", PandasUDFType.SCALAR)
def normPartitioner(x, y, z):
"""
"""
# Build vector of norms
norm = np.sqrt(x**2 + y**2 + z**2)
# Condition on the norm value
cond = 87
# Flag values based on condition
ret = np.ones_like(norm)
ret[norm <= cond] = 0
return pd.Series(ret)
df_colid = df.withColumn("partition_id", normPartitioner(df["x"], df["y"], df["z"]))
Going beyond: column partitioning as a standard Spark SQL method
Actually you do not even need to know or create the underlined partitioners. Say you already have a column that described your partitioning, then you can directly apply repartitionByCol:
from pyspark3d.repartitioning import repartitionByCol
df.show()
+---+----+
|Age|flag|
+---+----+
| 20| 0|
| 30| 1|
| 24| 0|
| 56| 0|
| 34| 1|
+---+----+
df_repart = repartitionByCol(df, "flag", preLabeled=True, numPartitions=2)
df_repart.show()
+---+----+
|Age|flag|
+---+----+
| 20| 0|
| 24| 0|
| 56| 0|
| 30| 1|
| 34| 1|
+---+----+
If your columns is not preLabeled (i.e. does not contain partition ID), this is not a problem:
from pyspark3d.repartitioning import repartitionByCol
df.show()
+---+----+----+
|Age|flag|part|
+---+----+----+
| 20| 0| A|
| 30| 2| A|
| 24| 0|toto|
| 56| 0| A|
| 34| 2|toto|
+---+----+----+
df_repart = repartitionByCol(df, "part", preLabeled=False)
df_repart.show()
+---+----+----+
|Age|flag|part|
+---+----+----+
| 20| 0| A|
| 30| 2| A|
| 56| 0| A|
| 24| 0|toto|
| 34| 2|toto|
+---+----+----+
df_repart.rdd.getNumPartitions()
2
Note however that the execution time will be longer since the routine needs to map column elements to distinct keys.
How is spark3D df.repartitionByCol different from Apache Spark df.repartitionByRange or df.orderBy?
df.repartitionByRangereturns aDataFramepartitioned by the specified column(s).df.orderByreturns aDataFramepartitioned and sorted by the specified column(s).
A major problem of both is there is no guarantee to always get the same final partitioning. In other words, they are non-deterministic. See for example SPARK-26024. For performance reasons they use sampling to estimate the ranges (with default size of 100). Hence, the output may not be consistent, since sampling can return different values.
Conversely df.repartitionByCol always guarantees the same final partitioning, by strictly following the partitioning scheme in the specified column. Obviously this exactness has a cost, and using df.repartitionByCol can increase the repartitioning time as shown in:
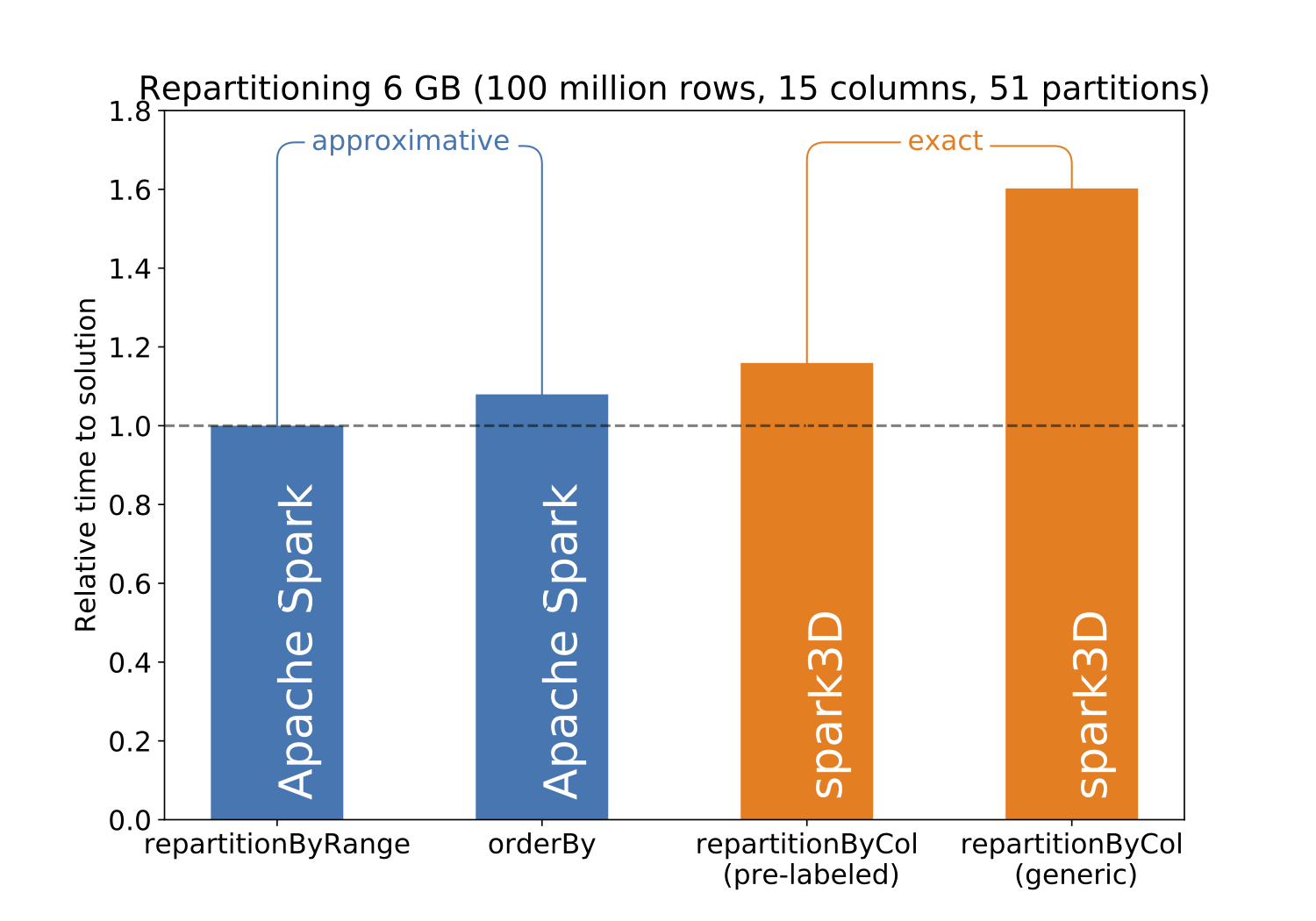
Note that the Spark df.repartition method is not present from this benchmark as for such volume of data it leads to completely unpredictive results (empty partitions, and partitions twice the size of others… even by fixing the number of partitions AND the column to use for the repartitioning).
Memory and speed considerations
We advice to cache the re-partitioned sets, to speed-up future call by not performing the re-partitioning again.
However keep in mind that if a large numPartitions decreases the cost of performing future queries (cross-match, KNN, …), it increases the partitioning cost as more partitions implies more data shuffle between partitions and often increases the scheduler delays. There is no magic number for numPartitions which applies in general, and you’ll need to set it according to the needs of your problem. But here are few guidelines:
What kind of performance I will get assuming fixed cluster configuration and input data set?
Assuming you use a fix cluster configuration (numbers of cores + available memory), the repartitioning time is quadratic in the final number of partitions:
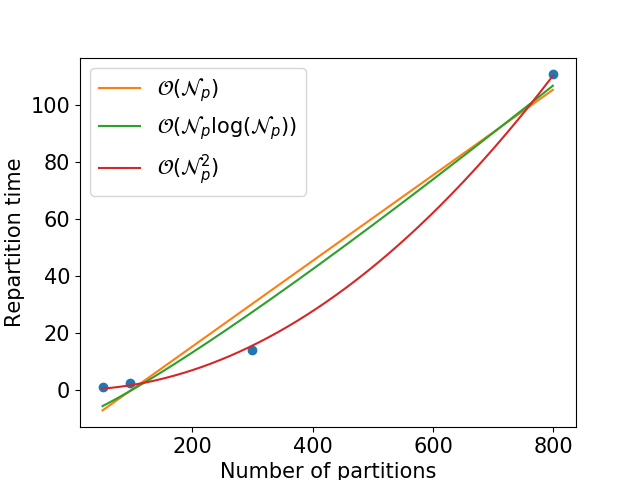
In fact, that also depends on the difference between the number of worker threads and the number of partitions:
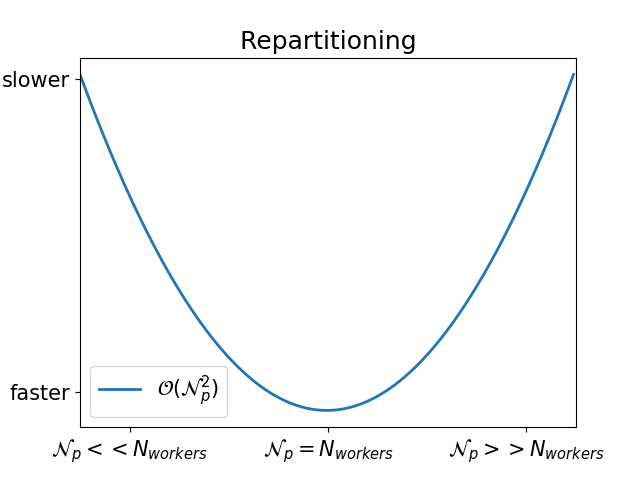
How can I check that my repartitioning is well balanced?
There is a method in spark3D to check the load balancing:
from pyspark3d.checkers import checkLoadBalancing
checkLoadBalancing(df_repart, "frac").show()
+--------+------------+
|Load (%)|partition_id|
+--------+------------+
| 10.52| 0|
| 10.19| 1|
| 9.925| 2|
| 9.87| 3|
| 10.135| 4|
| 10.13| 5|
| 9.49| 6|
| 9.81| 7|
| 9.87| 8|
| 10.06| 9|
+--------+------------+